Epithelial cell adhesion molecule
|
Read other articles:

This article relies largely or entirely on a single source. Relevant discussion may be found on the talk page. Please help improve this article by introducing citations to additional sources.Find sources: Abbotsford International Airshow – news · newspapers · books · scholar · JSTOR (August 2010) Abbotsford International AirshowThe Canadian Snowbirds on the Hotline at the Abbotsford Airshow 2000GenreAir showDatesAugustVenueAbbotsford International Airp...

Anan NurakhmanKomandan Korem 061/Surya KencanaPetahanaMulai menjabat 27 April 2023PendahuluRudy SaladinKomandan Korem 074/WarastratamaMasa jabatan29 Agustus 2022 – 27 April 2023PendahuluAchiruddinPenggantiAli AkhwanKomandan Grup A PaspampresMasa jabatan2021 – 29 Agustus 2022PendahuluAchiruddinPenggantiFaisol Izuddin Karimi Informasi pribadiLahir21 Desember 1976 (umur 46)Surakarta, Jawa TengahSuami/istriNy. Retno Setyas Tuti[1]Anak2Alma materAkademi Milit...

British actress (1926–2009) Zena MarshallBornZena Moyra Marshall(1926-01-01)1 January 1926Nairobi, KenyaDied10 July 2009(2009-07-10) (aged 83)London, EnglandNationalityBritishOccupationActressYears active1945–1967Spouses Paul Adam (m. 1947; div. 1953) Alexander Ward (m. 1967; div. 1969) Ivan Foxwell (m. 1991; died 2002) Zena ...

Eurovision Song Contest 2022Country UkraineNational selectionSelection processSelection among Vidbir 2022 participantsSelection date(s)22 February 2022Selected entrantKalush OrchestraSelected songStefaniaSelected songwriter(s)Ihor DidenchukIvan KlymenkoOleh PsiukTymofii MuzychukVitalii DuzhykFinals performanceSemi-final resultQualified (1st, 337 points)Final result1st, 631 pointsUkraine in the Eurovision Song Contest ◄2021 • 2022 • 2023► Ukraine...

「バースデー」はこの項目へ転送されています。その他の用法については「バースデイ」をご覧ください。 この記事は検証可能な参考文献や出典が全く示されていないか、不十分です。出典を追加して記事の信頼性向上にご協力ください。(このテンプレートの使い方)出典検索?: 誕生日 – ニュース · 書籍 · スカラー · CiNii · J-STAGE · ND...

Ayasse Un vieux pont sur l'Ayasse à Pontboset. Caractéristiques Longueur 23 km Bassin 110 km2 Bassin collecteur Pô Cours Source Lac Misérin · Localisation Vallée de Champorcher Confluence Doire Baltée · Localisation Hône Géographie Pays traversés Italie Régions traversées Vallée d'Aoste Principales localités Champorcher, Pontboset, Hône modifier L'Ayasse est un affluent de la Doire Baltée qui coule dans la Vallée de Champorcher, dans la basse Vallée d'Aoste. G

Feudal council of the Kingdom of Jerusalem This article includes a list of references, related reading, or external links, but its sources remain unclear because it lacks inline citations. Please help to improve this article by introducing more precise citations. (May 2017) (Learn how and when to remove this template message) Coat of arms of the Kingdom of Jerusalem. The Haute Cour (English: High Court) was the feudal council of the Kingdom of Jerusalem. It was sometimes also called the curia...

128-ма гвардійська гірськострілецька дивізія рос. 128-я гвардейская горнострелковая дивизияНа службі 12 липня 1922 — 15 грудня 1956Країна Радянський СоюзВид Червона арміяТип гірська піхотаВійни/битви Друга світова війнаУгорщина 1956Нагороди Почесні найменування «Туркеста

Pemandangan Sungai Citumang Sungai Citumang adalah sebuah sungai yang terletak di Desa Bojong, Kecamatan Parigi, Kabupaten Pangandaran, Provinsi Jawa Barat. Sungai ini berada di kawasan hutan milik Perhutani.[1] Sungai ini memiliki luas mencapai 6,6 hektar dengan suhu udara 25 °C-28 °C.[2] Toponimi Sungai tersebut dinamakan 'Citumang' dikarenakan aliran airnya berada di atas gua atau biasa disebut numpang, sedangkan ci diambil dari kata cai yang berarti air.[2&...

Church in Saint Petersburg, RussiaChurch of Our Lady of KazanRussian: Церковь Казанской иконы Божьей Матери Finnish: Terijoen ortodoksinen kirkko60°11′32″N 29°42′23″E / 60.192311°N 29.70627°E / 60.192311; 29.70627LocationZelenogorsk, Saint PetersburgCountryRussiaDenominationRussian OrthodoxHistoryConsecrated1915, destroyed 1939;reconsecrated 1990ArchitectureArchitect(s)Nikolay NikonovStyleRussianGroundbreaking1910Completed19...

Canadian medical organization This article relies excessively on references to primary sources. Please improve this article by adding secondary or tertiary sources. Find sources: Canadian Organ Replacement Registry – news · newspapers · books · scholar · JSTOR (November 2010) (Learn how and when to remove this template message) The Canadian Organ Replacement Registry (CORR) is a health organization started by Canadian nephrologists and kidney transplan...

Ship of the Baltic fleet, 1703–27 Shtandart Modern Replica (1999) under sail in Baltic Sea, 2007 History Russian Empire NameShtandart NamesakeImperial Yacht ship, frigate (Modern Replica, 1999) OwnerImperial Russian Navy Ordered1703 BuilderShipyard of Olonets city Laid downApril 24, 1703 LaunchedAugust 22, 1703 AcquiredSeptember 8, 1703 CommissionedSeptember 8, 1703, 1999 Decommissioned1727 In service1710 Out of service1711 FateBroken up General characteristics (typical) Class and type24-gu...
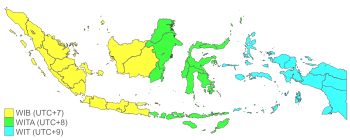
Halaman ini berisi artikel tentang salah satu zona waktu di Indonesia. Untuk acara televisi NET. yang bernama sama, lihat Waktu Indonesia Timur (acara televisi). Zona waktu IndonesiaPeta zona waktu Indonesia Waktu terkini Waktu Indonesia Barat (UTC+07:00)01:07, December 1, 2023 WIB [refresh]Waktu Indonesia Tengah (UTC+08:00)02:07, December 1, 2023 WITA [refresh]Waktu Indonesia Timur (UTC+09:00)03:07, December 1, 2023 WIT [refresh] Waktu Indonesia Timur (disingkat WIT) adalah salah satu dari t...

2013 South Korean filmNo BreathingPromotional poster for No BreathingHangul노브레싱Revised RomanizationNobeureshing Directed byJo Yong-sunWritten byYoo Young-ah Jo Yong-sunProduced byLee Song-joon Lee Geun-wook Park Chang-hyun Jung Dae-hoon Park Se-joon Park Do-wonStarringLee Jong-suk Seo In-guk Kwon YuriCinematographyLee Joon-gyuEdited byHeo Sun-miMusic byMarcoProductioncompanies9ers EntertainmentSoo Jack FilmsUnion Investment PartnersDistributed byCineguru/Daou TechRelease date October&...

SorabeJenis aksara Abjad BahasaMalagasiPeriodeabad ke-17 hingga 1823Aksara terkaitSilsilahHieroglif MesirProto-SinaiFenisiaAramNabathArabPegon? (diperdebatkan)[1][2]Sorabe Artikel ini mengandung transkripsi fonetik dalam Alfabet Fonetik Internasional (IPA). Untuk bantuan dalam membaca simbol IPA, lihat Bantuan:IPA. Untuk penjelasan perbedaan [ ], / / dan ⟨ ⟩, Lihat IPA § Tanda kurung dan delimitasi transkripsi. Abjad Sorabe (juga...

Square in Dublin, Ireland Ormond SquareOrmond Square in 2012Native nameCearnóg Urumhan (Irish)NamesakeJames Butler, first Duke of OrmondeLocationDublin, IrelandPostal codeD07Coordinates53°20′47″N 6°16′14″W / 53.3463°N 6.2705°W / 53.3463; -6.2705 Ormond Square (Irish: Cearnóg Urumhan)[1] is a square on the northside of Dublin city. History Ormond Square sits on the site of the former Ormond Market. Along with Ormond Quay, the square is named a...

Bridge in ShrewsburyKingsland BridgeKingsland BridgeCoordinates52°42′16″N 2°45′32″W / 52.7044°N 2.7589°W / 52.7044; -2.7589OS grid referenceSJ488121CrossesRiver SevernLocaleShrewsburyHeritage statusGrade IIPreceded byPorthill BridgeFollowed byGreyfriars BridgeCharacteristicsDesignarchLongest span212 ft (65 m)HistoryDesignerJohn William GroverConstructed byCleveland Bridge & Engineering CompanyConstruction start1883Construction end1883Statistic...

Educational publisher McGraw Hill redirects here. For the business and financial information company previously known as McGraw Hill Financial, see S&P Global. McGraw HillThe branded McGraw Hill logo as of 2020[update]Founded1888; 135 years ago (1888)FounderJames H. McGrawJohn A. HillCountry of originUnited StatesHeadquarters locationNew York CityKey peopleSimon AllenPublication typesAdaptive learning technology, educational software, e-books, apps, platform serv...

1998 unique amphibious assault ship of the Royal Navy For other ships with the same name, see HMS Ocean. HMS Ocean during Operation Ellamy and the 2011 military intervention in Libya History United Kingdom NameHMS Ocean OperatorRoyal Navy Ordered11 May 1993 BuilderVickers Shipbuilding and Engineering Ltd, Kværner (Govan) Laid down30 May 1994 Launched11 October 1995 Sponsored byQueen Elizabeth II of the United Kingdom Commissioned30 September 1998 Decommissioned27 March 2018[1] RefitM...

Балка Крутенька(заповідне урочище) 47°42′28″ пн. ш. 34°44′00″ сх. д. / 47.70777800002777269° пн. ш. 34.7336110000277784593° сх. д. / 47.70777800002777269; 34.7336110000277784593Координати: 47°42′28″ пн. ш. 34°44′00″ сх. д. / 47.70777800002777269° пн. ш. 34.7336110000277784593° сх. д.þ...






